Structural Imaging Approaches Shed Important Light on Novel Coronavirus Entry and Infectivity
Unique Features May be Critical in the Development of Vaccines and Therapeutics
With the sudden emergence of the novel coronavirus, SARS-CoV-2 the causative agent of COVID-19, scientists rapidly mobilized to map the route of infection and the underlying mechanism of cell entry. The race was on to characterize the coat proteins from this novel coronavirus and decipher the molecular interactions with host cells.
In the wake of the previous SARS-CoV virus, which also caused acute respiratory disease, scientists found that upon entry into the lung tissue, the virus gained host-cell access through the interaction of an outer virus coat protein (CoV spike (S) glycoprotein) with angiotensin-converting enzyme 2 (ACE2) receptors on the surface of host cells. Subsequent research identified the CoV S protein as a key virulence factor and a target for vaccine, therapeutic antibodies, and diagnostics.
As shown in a recent study, the 2019-CoV S protein (from the novel SARS-CoV-2 virus responsible for the current outbreak) is shown to serve a central role in mediating virus entry as well. Coincidentally, this mechanism also involves host ACE2 receptor binding. The question immediately surfaced, were the molecular details similar to the previous mechanism and could this 2019-CoV S protein once again be targeted for therapeutic development? Detailed high-resolution imaging of the protein has revealed both expected and novel structural elements, uncovering unique capabilities previously not observed in this family of coronaviruses.
The Use of Cryo-Electron Microscopy to Image the 2019-CoV Spike Glycoprotein
The study, “Cryo-EM Structure of the 2019-nCoV spike in the prefusion conformation”, involved a collaboration of scientists from University of Austin, TX and the National Institute of Allergy and Infectious Diseases, who teamed up to perform structural and biophysical analysis of the 2019-CoV S protein complex from the novel virus. In its native state, 2019-CoV S is a densely glycosylated membrane protein arranged in a complex of three protein subunits. This trimer adjusts its conformation upon receptor interaction thus mediating the entry process.
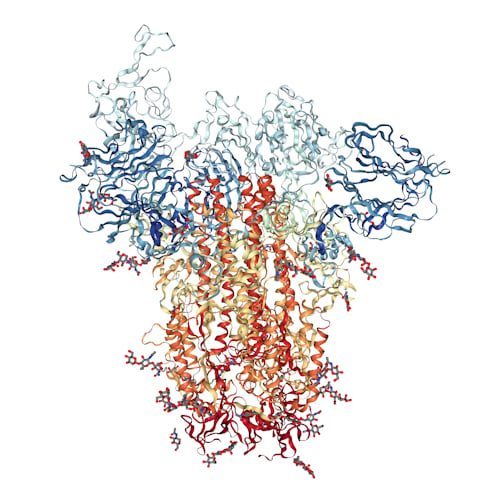
Due to the inherent limitations in expressing, purifying, and imaging membrane proteins, the ecto-domain portion of the protein, which faces the exterior of the virus and constitutes the receptor binding site, was targeted. First, the n-terminal ectodomain residues 1 to 1208 from 2019-nCoV S protein was modified with the addition of two stabilizing proline residues in the C-terminal end, a strategy that worked well in previous investigations of coronavirus S proteins. The protein was next expressed in recombinant cells and purified by affinity and size-exclusion chromatography techniques. Purified protein was then used in preparation of Cryo-EM (cryo electron microscopy) grids, within which the fully glycosylated protein adopted high density arrays with minimal aggregation.
Cryo-EM data collection began with imaging the liquid ethane treated frozen grid by use of a Titan Krios EM instrument fitted with a K3 detector. Movies were collected at x22,500 magnification corresponding to a calibrated pixel size of 1.047 Å/pixel. Computational software was used to pick appropriate particles which were collated and subjected to two dimensional, three dimensional, and non-uniform refinement. The image maps were sharpened and the protein sequences were used to fit the electron density of the structures.
(Shown is the cryo-EM structure of the SARS-CoV S protein complex -- publication and image referenced below)
Structure of the 2019-CoV S Protein
Upon collecting and processing 3207 micrograph movies, the investigators arrived at a 3.5 Å-resolution three-dimensional reconstruction of the trimeric protein complex. The asymmetric structure contained one receptor-binding domain (RBD) in an “up” conformation, with the other two in the “down” position.
In the current context, the cryo-EM technique excelled at high-resolution structure determination where others techniques may have been challenged due to the complex nature of the protein and the conditions. Structural determination by x-ray crystallography, for instance, requires highly uniformed crystallization of proteins. Such a structure would be limited to a single individual 2019-nCoV S subunit captured in a static conformation owing to requirements for symmetry in the crystal lattice. Any movement or breathing of the molecule would significantly interfere or be unresolved upon crystallization and subsequent analysis. Furthermore, modifications including glycosylation make the overall analysis even more tedious. Cryo EM was the right technique to use -- providing the highest-quality information on the protein complex, rapidly and conclusively.
Similarities with Previous Coronavirus Cov S Proteins
The powerful cryo-EM imaging approach disclosed several similarities between 2019-CoV S and previous CoV S proteins. First off, the 2019-CoV S backbone structure is very similar to SARS-CoV S with a root mean square deviation of 3.8 Å over 959 Cα atoms. There is a high level of homology between the individual structural domains. The two share 98% sequence identity with the most notable variation coming from an amino acid insertion in a protease cleavage site, resulting in a RRAR furin recognition sequence unique from SARS-CoV S.
Critical Differences between the 2019-CoV S and SARS-CoV S Proteins
The difference in 2019-CoV S and the creation of a polybasic furin protease site, like that mentioned above, is a hallmark of highly virulent proteins, such as hemagglutinin protein from avian and human influenza viruses.
Another key difference between the proteins involves the position of the down conformations. The receptor binding site (RBD) of SARS-CoV packs tightly against the N-terminal domain while the 2019-CoV RBD is angled more towards the central cavity of the complex.
Translation of Differences to Virulence and Infectivity
The fact that the proteins were similar in sequence and structure, and bound to the same host protein receptor angiotensin-converting enzyme 2 (ACE2), prompted the investigators to analyze the kinetics of receptor binding.
Surface Plasmon Resonance experiments demonstrated the 2019-CoV S protein bound with very high (15 nM) affinity. Interestingly, this was over 10-fold more tightly than the SARS-CoV S protein with the same protein receptor. The overall structures of both bound complexes were confirmed by negative stain EM.
Structural and Functional Implications for Disease
As is the case with many emerging pathogens, it is the differences – however slight they may be, that can make the distinction between novel diseases and previously seen outbreaks.
With regard to the 2019-CoV S protein in comparison with the previous SARS-CoV S protein, the authors suggest that the large increase in binding strength of 2019-CoV S with its ACE2 receptor may translate to greater propensity for cell entry and higher infectivity rate of the virus. Of course, this argument lies beyond the limitations of structural analysis and in the domain of physiological investigations, as the authors also state. Nonetheless, high-resolution approaches, such as cryo-EM used here, can disclose fine-structural elements that can provide clues to important functional differences.
Amidst the SARS outbreak in 2002, monoclonal antibodies against the SARS-CoV RBD were created in the lab and used in research and therapeutic design efforts. The similarities between the SARS-CoV S and the 2019-CoV S proteins shown in this study justly led the investigators to measure cross-reactivity of the two with these monoclonal antibodies.
Despite the strong structural similarities, 2019-CoV S showed minimal binding activity to the antibodies, even at concentrations favorable to SARS-CoV antibody binding. The results demonstrate that distinct antibodies or antibody sources must be investigated and used in research and therapeutics discovery work. Nonetheless, this is another example of the effects of subtle alterations or mutations and larger biological implications – details that may be exploited in therapeutic and preventative development.
Summary and Outlook
Several high-profile published and ongoing studies are looking at the effects of protease inhibitors and antibodies in blockade of the 2019-CoV S protein mediated cell entry. These include use of clinically approved protease inhibitors and antibodies derived from sera from convalescent SARS patients. Both show promising signs of (cross)reactivity and potential as treatment candidates. Many other therapeutic prospects and vaccine candidates are in the pipeline as well.
The tenacity and urgency with which scientists and health care professionals have approached this rapidly emerging problem is impressive to say the very least. LitCovid, a curated literature hub created by the NIH National Library of Medicine for tracking up-to-date publications on the novel coronavirus, currently houses 2018 published studies – with the addition of nearly 500 in the week spanning March 23 to 29 alone. This number continues to grow by the hour and the day.
Todays arsenal of innovation and advanced technology is actively engaging this unprecedented outbreak. In the end, new levels of scientific and medical firepower will surely meet the challenge and prevail.
Image courtesy of the Protein Data Bank
https://www.rcsb.org/3d-view/6CRZ
Kirchdoerfer, R.N., Wang, N., Pallesen, J. et al. Stabilized coronavirus spikes are resistant to conformational changes induced by receptor recognition or proteolysis. (2018) Sci Rep, 8: 15701. doi: 10.1038/s41598-018-34171-7.
H.M. Berman, J. Westbrook, Z. Feng, G. Gilliland, T.N. Bhat, H. Weissig, I.N. Shindyalov, P.E. Bourne.
(2000) The Protein Data Bank Nucleic Acids Research, 28: 235-242. doi: 10.1093/nar/28.1.235.
RCSB Protein Data Bank: biological macromolecular structures enabling research and education in fundamental biology, biomedicine, biotechnology and energy. (2019) Nucleic Acids Research 47: D464–D474. doi: 10.1093/nar/gky1004.