Real-Time vs. Endpoint PCR: Choosing the Right Method for Your Experiment
GEMINI (2025)
The Polymerase Chain Reaction (PCR) is the cornerstone of molecular biology, offering laboratory professionals the indispensable capability to amplify specific deoxyribonucleic acid (DNA) sequences from minute samples. The proper selection between a Real-Time PCR (
Defining the Fundamentals of Polymerase Chain Reaction Assays
The Polymerase Chain Reaction, irrespective of its format, relies on fundamental thermocycling principles: denaturation, annealing, and extension. The key distinction between Real-Time and conventional methodologies lies in the phase of the amplification process where data is collected, fundamentally altering the nature of the resulting data—quantitative versus qualitative.
What is Endpoint PCR?
Endpoint PCR, or conventional PCR, is a foundational molecular technique employed primarily for the qualitative detection of a target DNA sequence. The reaction is performed in a standard thermal cycler, undergoing 25 to 40 cycles of amplification. Crucially, the data is collected only after the amplification phase is complete—at the endpoint of the reaction.
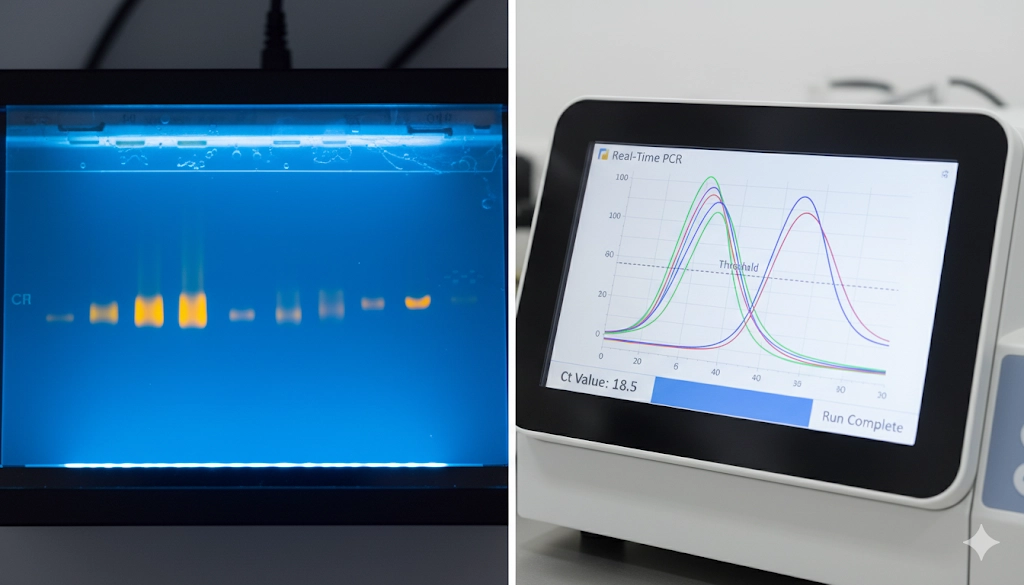
Upon completion of the thermocycling, the amplified DNA products, or amplicons, are visualized and analyzed using gel electrophoresis . The presence of a band of the expected size confirms the initial presence of the target sequence in the sample, while the intensity of the band provides only a qualitative or semi-quantitative estimation of the final product yield. Due to the endpoint measurement, this method inherently measures the quantity of product in the plateau phase, where reagents become limiting and the reaction efficiency decreases, making it unreliable for precise quantification.
What is Real-Time PCR (qPCR)?
Real-Time PCR (
The
Kinetic Measurement and the Threshold Cycle (Ct ) Value
The most significant functional difference between the two techniques is the point of measurement, which dictates the ability to perform accurate absolute and relative quantification. Endpoint PCR detects the final yield, whereas
The Exponential Phase and Quantitative Precision
In an ideal PCR reaction, the amount of product should double during each cycle in the exponential phase. Real-Time PCR captures this linear relationship between the initial template concentration and the cycle number required to reach a detectable signal. This is defined by the threshold cycle (
A key principle is the inverse relationship between the initial target concentration and the
The Plateau Phase Limitation in Endpoint PCR
Endpoint PCR, by its nature, provides measurements during the plateau phase. The plateau is reached when reaction components—such as deoxyribonucleotides (dNTPs), primers, or enzyme activity—become limiting, or when product inhibition occurs. Even if two samples began with vastly different initial target concentrations, they may yield similar amounts of product when the reaction is arrested at the plateau phase, completely masking any initial quantitative differences.
This limitation means that while conventional PCR is excellent for confirming the presence of a gene, any attempt to use band intensity for accurate comparison or quantification of starting material introduces significant error and lack of reproducibility, making it an unsuitable technique for demanding molecular assays that require high quantitative accuracy.
Methodology, Instrumentation, and Assay Design Considerations
The methodological workflow and required instrumentation vary substantially, impacting both the initial setup cost and the post-amplification workload for laboratory professionals.
Feature | Endpoint PCR (Conventional) | Real-Time PCR ( |
|---|---|---|
Measurement Timing | Post-amplification (Endpoint/Plateau Phase) | During amplification (Real-Time/Exponential Phase) |
Data Type | Qualitative (Presence/Absence) or Semi-Quantitative | Absolute or Relative Quantitative |
Detection Method | Agarose Gel Electrophoresis and Staining | Fluorescent Dyes (e.g., SYBR Green) or Probes (e.g., TaqMan) |
Instrumentation | Standard Thermal Cycler | Thermal Cycler with Integrated Fluorometer |
Post-Reaction Workload | High (Gel casting, running, imaging, waste disposal) | Low (Data analyzed immediately by software) |
Contamination Risk | High (Sample handling post-amplification) | Low (Closed-tube system) |
The choice of detection chemistry further differentiates the Real-Time PCR assay. Intercalating fluorescent dyes, such as SYBR Green, bind to double-stranded DNA and are cost-effective but non-specific, potentially detecting primer dimers. Alternatively, sequence-specific fluorescent probes, such as TaqMan probes, offer superior specificity by only generating a signal upon hydrolysis by the Taq polymerase, confirming that fluorescence is derived exclusively from the target amplicon. This specificity is often critical for complex diagnostic and research assays.
Strategic Application and Experimental Context
The selection between Endpoint PCR and Real-Time PCR should be driven by the specific experimental question posed, the required level of quantification, and resource availability.
Applications for Highly Quantitative Real-Time PCR
Real-Time PCR is the gold standard when precise quantification and high sensitivity are mandatory. Key applications include:
Gene Expression Analysis: Accurately measuring the relative expression levels of messenger RNA (mRNA) transcripts between different samples, typically using a housekeeping gene as a reference.
Viral Load Determination: Precise measurement of the number of viral genomes (e.g., copies/mL) in a clinical sample, crucial for monitoring infectious disease progression and treatment efficacy.
Copy Number Variation (CNV) Analysis: Determining the number of copies of a specific gene region in a genome.
High-Throughput Screening: The reduced post-amplification steps and reliance on automated data analysis make
qPCR assays highly scalable.
Appropriate Uses for Conventional Endpoint PCR
Despite the quantitative superiority of
Qualitative Detection: Simple presence/absence confirmation of a gene or pathogen, such as screening bacterial colonies for a specific plasmid insertion.
Genotyping and Allele Detection: Used to amplify specific markers followed by restriction enzyme digestion or sequencing for genomic analysis.
Product Preparation for Downstream Applications: Generating sufficient amounts of DNA for use in cloning, sequencing preparation, or other molecular biology techniques.
Low-Cost Screening: When budgetary constraints are strict and only a yes/no answer regarding target presence is required, conventional PCR remains a viable option.
Optimal Selection for Molecular Quantification
The decision tree for molecular detection and quantification must prioritize data integrity and experimental objective. While Endpoint PCR offers rapid, low-cost qualitative results suitable for cloning or simple presence testing, Real-Time PCR assays provide the kinetic data necessary for highly accurate and reproducible quantification of nucleic acid targets. Laboratory professionals should select
Frequently Asked Questions (FAQ)
What is the primary advantage of Real-Time PCR over conventional PCR for viral load assays?
The primary advantage is the ability of Real-Time PCR (
Can Endpoint PCR be used for reliable gene expression quantification?
No. Endpoint PCR measures product after the reaction has reached the plateau phase, where reagents are limiting and reaction efficiency is variable. This makes the final product yield an inaccurate reflection of the initial template concentration, rendering it unsuitable for reliable gene expression quantification.
What detection chemistries are commonly used in a quantitative Real-Time PCR assay?
The most common detection chemistries are non-specific intercalating dyes (e.g., SYBR Green), which bind all double-stranded DNA, and sequence-specific probes (e.g., TaqMan probes), which use fluorescence resonance energy transfer (FRET) for highly specific amplicon detection.
What is the meaning of the Ct value in Real-Time PCR analysis?
The
This article was created with the assistance of Generative AI and has undergone editorial review before publishing.